AutoGRAPH was an integrated
web server for multi-species comparative genomic analysis. It
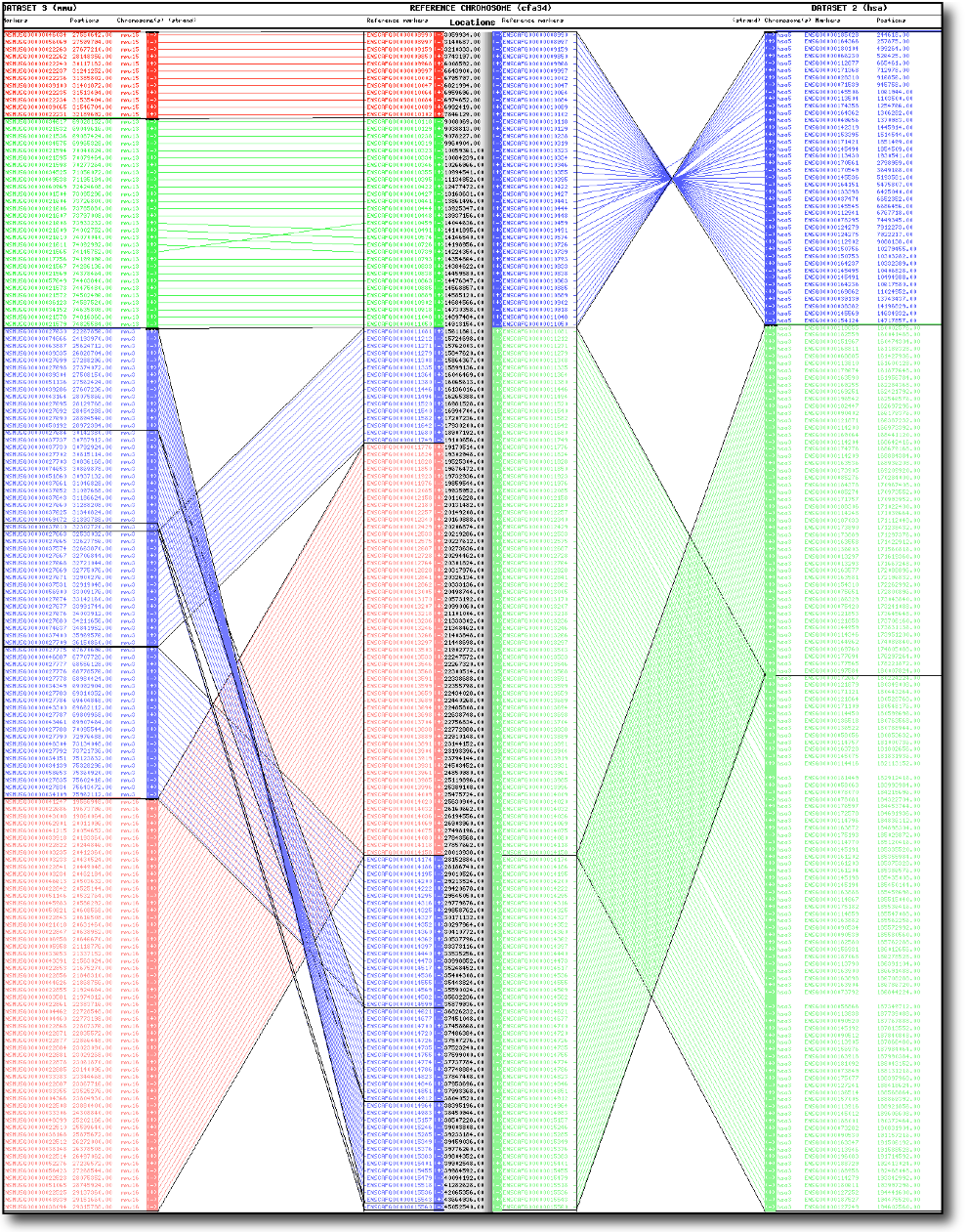
is designed for constructing and visualizing synteny
maps between two or three species, determination and display of
macrosynteny and microsynteny relationships among species, and
for highlighting evolutionary breakpoints.
The web server constructs synteny maps by pairwise comparison of marker/anchor orders between
a reference chromosome and one or two tested genome(s).
It permits users to visualize and characterize several features:
Conserved segments (CS)
, Conserved Segments Ordered (CSO)
, breakpoints
, marker density in these regions, inferred locations for 1:0 orthologous relationships...
AutoGRAPH is a versatile tool and can be utilized for the integration and comparison of different maps (meiotic, RH...)
with sequence resources within a single species. |